The following publications have used the Uni-Mol software. Publications that only mentioned the Uni-Mol will not be included below.
We encourage explicitly mentioning Uni-Mol with proper citations in your publications, so we can more easily find and list these publications.
Last update date: 11/21/2024
2024
Node-Aligned Graph-to-Graph: Elevating Template-free Deep Learning Approaches in Single-Step Retrosynthesis
Lin Yao, Wentao Guo, Zhen Wang, Shang Xiang, Wentan Liu, Guolin Ke
Jacs Au, 2024, 4 (3), 992–1003.
DOI: 10.1021/jacsau.3c00737
Learning Multi-view Molecular Representations with Structured and Unstructured Knowledge
Yizhen Luo, Kai Yang, Massimo Hong, Xing Yi Liu, Zikun Nie, Hao Zhou, Zaiqing Nie
arXiv, 2024, 2406.09841.
DOI: 10.48550/arXiv.2406.09841
Mol-AE: Auto-Encoder Based Molecular Representation Learning With 3D Cloze Test Objective
Junwei Yang, Kangjie Zheng, Siyu Long, Zaiqing Nie, Ming Zhang, Xinyu Dai, Wei-Ying Ma, Hao Zhou
Biorxiv, 2024.
DOI: 10.1101/2024.04.13.589331
MoleculeCLA: Rethinking Molecular Benchmark via Computational Ligand- Target Binding Analysis
Shikun Feng, Jiaxin Zheng, Yinjun Jia, Yanwen Huang, Fengfeng Zhou, Wei-Ying Ma, Yanyan Lan
arXiv, 2024, 2406.17797.
DOI: 10.48550/arXiv.2406.17797
Towards 3D Molecule-Text Interpretation in Language Models
Sihang Li, Zhiyuan Liu, Yanchen Luo, Xiang Wang, Xiangnan He, Kenji Kawaguchi, Tat-Seng Chua, Qi Tian
arXiv, 2024, 2401.13923.
DOI: 10.48550/arXiv.2401.13923
3D-MolT5: Towards Unified 3D Molecule-Text Modeling with 3D Molecular Tokenization
Qizhi Pei, Lijun Wu, Kaiyuan Gao, Jinhua Zhu, Rui Yan
arXiv, 2024, 2406.05797.
DOI: 10.48550/arXiv.2406.05797
From Theory to Therapy: Reframing SBDD Model Evaluation via Practical Metrics
Bowen Gao, Haichuan Tan, Yanwen Huang, Minsi Ren, Xiao Huang, Wei-Ying Ma, Ya-Qin Zhang, Yanyan Lan
arXiv, 2024, 2406.08980.
DOI: 10.48550/arXiv.2406.08980
MolBind: Multimodal Alignment of Language, Molecules, and Proteins
Teng Xiao, Chao Cui, Huaisheng Zhu, Vasant G. Honavar
arXiv, 2024, 2403.08167.
DOI: 10.48550/arXiv.2403.08167
MocFormer: A Two-Stage Pre-training-Driven Transformer for Drug-Target Interactions Prediction
Yi-Lun Zhang, Wen-Tao Wang, Jia-Hui Guan, Deepak Kumar Jain, Tian-Yang Wang, Swalpa Kumar Roy
Int J Comput. Intell Syst, 2024, 17 (1), 165.
DOI: 10.1007/s44196-024-00561-1
Protein-ligand binding representation learning from fine-grained interactions
Shikun Feng, Minghao Li, Yinjun Jia, Weiying Ma, Yanyan Lan
arXiv, 2023, 2311.16160.
DOI: 10.48550/arXiv.2311.16160
ProFSA: Self-supervised Pocket Pretraining via Protein Fragment- Surroundings Alignment
Bowen Gao, Yinjun Jia, Yuanle Mo, Yuyan Ni, Weiying Ma, Zhiming Ma, Yanyan Lan
arXiv, 2024, 2310.07229.
DOI: 10.48550/arXiv.2310.07229
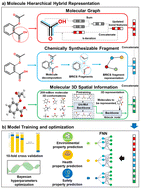
An interpretable 3D multi-hierarchical representation-based deep neural network for environmental, health and safety properties prediction of organic solvents
Jun Zhang, Qin Wang, Yang Lei, Weifeng Shen
Green Chem., 2024, 26 (7), 4181–4191.
DOI: 10.1039/D3GC04801B
Stereochemically-aware bioactivity descriptors for uncharacterized chemical compounds
Arnau Comajuncosa-Creus, Aksel Lenes, Miguel S'anchez- Palomino, Dylan Dalton, Patrick Aloy
J. Cheminformatics, 2024, 16 (1), 70.
DOI: 10.1186/s13321-024-00867-4
DeepP450: Predicting Human P450 Activities of Small Molecules by Integrating Pretrained Protein Language Model and Molecular Representation
Jiamin Chang, Xiaoyu Fan, Boxue Tian
J. Chem. Inf. Model., 2024, 64 (8), 3149–3160.
DOI: 10.1021/acs.jcim.4c00115